Getting Started in R
The term “R” is used to refer to both the programming language and the software that interprets scripts written using it. RStudio is a popular way to not only write your R scripts but also to interact with the R software. To function correctly, RStudio needs R and therefore both need to be installed on your computer.
You may be asking yourself - why should I learn R and not use a program like Excel to analyze my amplicon data and perform statistical analyses? There are a couple reasons R is generally a useful program and is really beneficial for using in 16S amplicon analyses:
- R is great for reproducibility: R doesn’t involve a lot of pointing and clicking, which is good because you have scripts instead to make it clear what you did instead of remembering a series of buttons you pushed. Your code can be shared with collaborators or people interested in your work in the future and they can easily reproduce your analyses.
- R is interdisciplinary and extensive: There are 10,000+ packages that can be installed to extend the base R capabilities to apply statistical analyses and make graphics for image analysis, time series, genomics, and amplicon analyses, to name a couple of examples.
- R produces high-quality graphics: Plotting functions are endless, and allow you to easily adjust any aspect of your figures
- R is a free, open-source platform: More transparency, and constant updates to improve the user experience
- R is a large and welcoming community: Thousands of people use R on a daily basis, and there are many ways to get help through the RStudio Blog, Stack Overflow, or google searches.
RStudio Interface
Let’s start by learning about RStudio, which is an Integrated Development Environment (IDE) for working with R. The RStudio IDE open-source product is free under the Affero General Public License (AGPL) v3. The RStudio IDE is also available with a commercial license and priority email support from RStudio, Inc.
We will use RStudio IDE to write code, navigate the files on our computer, inspect the variables we are going to create, and visualize the plots we will generate. RStudio can also be used for other things (e.g., version control, developing packages, writing Shiny apps) that we will not cover during the workshop.
RStudio is divided into 4 “Panes”: the Source for your scripts and documents (top-left, in the default layout), your Environment/History (top-right), your Files/Plots/Packages/Help/Viewer (bottom-right), and the R Console (bottom-left). The placement of these panes and their content can be customized (see menu, Tools -> Global Options -> Pane Layout).
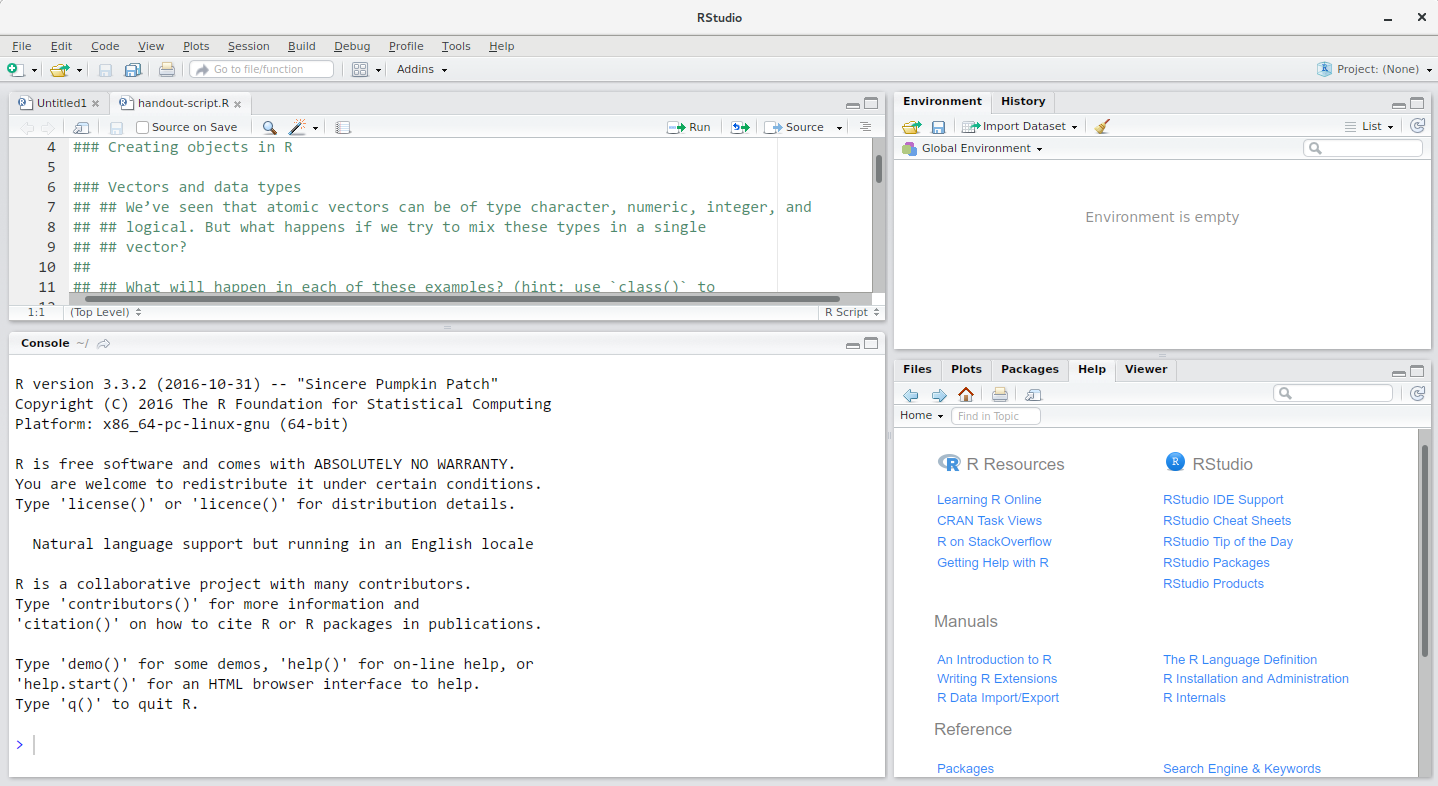
One of the advantages of using RStudio is that all the information you need to write code is available in a single window. Additionally, with many shortcuts, autocompletion, and highlighting for the major file types you use while developing in R, RStudio will make typing easier and less error-prone.
Getting set up
It is good practice to keep a set of related data, analyses, and text self-contained in a single folder, called the working directory. All of the scripts within this folder can then use relative paths to files that indicate where inside the project a file is located (as opposed to absolute paths, which point to where a file is on a specific computer). Working this way makes it a lot easier to move your project around on your computer and share it with others without worrying about whether or not the underlying scripts will still work.
RStudio provides a helpful set of tools to do this through its “Projects” interface, which not only creates a working directory for you, but also remembers its location (allowing you to quickly navigate to it) and optionally preserves custom settings and open files to make it easier to resume work after a break. Go through the steps for creating an “R Project” for this tutorial below.
- Start RStudio.
- Under the
Filemenu, click onNew Project. ChooseNew Directory, thenNew Project. - Enter a name for this new folder (or “directory”), and choose a convenient location for it. This will be your working directory for the rest of the day (e.g.,
~/amplicon-analysis). - Click on
Create Project. - (Optional) Set Preferences to ‘Never’ save workspace in RStudio.
RStudio’s default preferences generally work well, but saving a workspace to .RData can be cumbersome, especially if you are working with larger datasets. To turn that off, go to Tools –> ‘Global Options’ and select the ‘Never’ option for ‘Save workspace to .RData’ on exit.’
Organizing your Working Directory
Using a consistent folder structure across your projects will help keep things organized, and will also make it easy to find/file things in the future. This can be especially helpful when you have multiple projects. In general, you may create directories (folders) for scripts, data, and documents.
data_raw/&data/Use these folders to store raw data and intermediate datasets you may create for the need of a particular analysis. For the sake of transparency and provenance, you should always keep a copy of your raw data accessible and do as much of your data cleanup and preprocessing programmatically (i.e., with scripts, rather than manually) as possible. Separating raw data from processed data is also a good idea. For example, you could have filesdata_raw/tree_survey.plot1.txtand...plot2.txtkept separate from adata/tree.survey.csvfile generated by thescripts/01.preprocess.tree_survey.Rscript.documents/This would be a place to keep outlines, drafts, and other text.scripts/This would be the location to keep your R scripts for different analyses or plotting, and potentially a separate folder for your functions (more on that later).- Additional (sub)directories depending on your project needs.
For this workshop, we will need a data_raw/ folder to store our raw data, and we will use data/ for when we learn how to export data as CSV files, and a fig/ folder for the figures that we will save.
- Under the
Filestab on the right of the screen, click onNew Folderand create a folder nameddata_rawwithin your newly created working directory (e.g.,~/data-carpentry/). (Alternatively, typedir.create("data_raw")at your R console.) Repeat these operations to create adataand afigfolder.
We are going to keep the script in the root of our working directory because we are only going to use one file and it will make things easier.
The Working Directory
The working directory is an important concept to understand. It is the place from where R will be looking for and saving the files. When you write code for your project, it should refer to files in relation to the root of your working directory and only need files within this structure.
Using RStudio projects makes this easy and ensures that your working directory is set properly. If you need to check it, you can use getwd(). If for some reason your working directory is not what it should be, you can change it in the RStudio interface by navigating in the file browser where your working directory should be, and clicking on the blue gear icon “More”, and select “Set As Working Directory”. Alternatively you can use setwd("/path/to/working/directory") to reset your working directory. However, your scripts should not include this line because it will fail on someone else’s computer.
Interacting with R
The basis of programming is that we write down instructions for the computer to follow, and then we tell the computer to follow those instructions. We write, or code, instructions in R because it is a common language that both the computer and we can understand. We call the instructions commands and we tell the computer to follow the instructions by executing (also called running) those commands.
There are two main ways of interacting with R: by using the console or by using script files (plain text files that contain your code). The console pane (in RStudio, the bottom left panel) is the place where commands written in the R language can be typed and executed immediately by the computer. It is also where the results will be shown for commands that have been executed. You can type commands directly into the console and press Enter to execute those commands, but they will be forgotten when you close the session.
Because we want our code and workflow to be reproducible, it is better to type the commands we want in the script editor, and save the script. This way, there is a complete record of what we did, and anyone (including our future selves!) can easily replicate the results on their computer.
RStudio allows you to execute commands directly from the script editor by using the Ctrl + Enter shortcut (on Macs, Cmd + Return will work, too). The command on the current line in the script (indicated by the cursor) or all of the commands in the currently selected text will be sent to the console and executed when you press Ctrl + Enter. You can find other keyboard shortcuts in this RStudio cheatsheet about the RStudio IDE.
At some point in your analysis you may want to check the content of a variable or the structure of an object, without necessarily keeping a record of it in your script. You can type these commands and execute them directly in the console. RStudio provides the Ctrl + 1 and Ctrl + 2 shortcuts allow you to jump between the script and the console panes.
If R is ready to accept commands, the R console shows a > prompt. If it receives a command (by typing, copy-pasting or sent from the script editor using Ctrl + Enter), R will try to execute it, and when ready, will show the results and come back with a new > prompt to wait for new commands.
If R is still waiting for you to enter more data because it isn’t complete yet, the console will show a + prompt. It means that you haven’t finished entering a complete command. This is because you have not ‘closed’ a parenthesis or quotation, i.e. you don’t have the same number of left-parentheses as right-parentheses, or the same number of opening and closing quotation marks. When this happens, and you thought you finished typing your command, click inside the console window and press Esc; this will cancel the incomplete command and return you to the > prompt.
Introduction to R
Learning Objectives
- Define the following terms as they relate to R: object, assign, call, function, arguments, options.
- Assign values to objects in R.
- Learn how to name objects
- Use comments to inform script.
- Solve simple arithmetic operations in R.
- Call functions and use arguments to change their default options.
- Inspect the content of vectors and manipulate their content.
- Subset and extract values from vectors.
- Analyze vectors with missing data.
Creating Objects in R
You can get output from R simply by typing math in the console:
3 + 5## [1] 812 / 7## [1] 1.714286However, to do useful and interesting things, we need to assign values to objects. To create an object, we need to give it a name followed by the assignment operator <-, and the value we want to give it:
weight_kg <- 55<- is the assignment operator. It assigns values on the right to objects on the left. So, after executing x <- 3, the value of x is 3. The arrow can be read as 3 goes into x. For historical reasons, you can also use = for assignments, but not in every context. Because of the slight differences in syntax, it is good practice to always use <- for assignments.
In RStudio, typing Alt + - (push Alt at the same time as the - key) will write <- in a single keystroke in a PC, while typing Option + - (push Option at the same time as the - key) does the same in a Mac.
Objects can be given any name such as x, current_temperature, or subject_id. You want your object names to be explicit and not too long. They cannot start with a number (2x is not valid, but x2 is). R is case sensitive (e.g., weight_kg is different from Weight_kg). There are some names that cannot be used because they are the names of fundamental functions in R (e.g., if, else, for, see here for a complete list). In general, even if it’s allowed, it’s best to not use other function names (e.g., c, T, mean, data, df, weights). If in doubt, check the help to see if the name is already in use. It’s also best to avoid dots (.) within names. Many function names in R itself have them and dots also have a special meaning (methods) in R and other programming languages. To avoid confusion, don’t include dots in names. It is also recommended to use nouns for object names, and verbs for function names. It’s important to be consistent in the styling of your code (where you put spaces, how you name objects, etc.). Using a consistent coding style makes your code clearer to read for your future self and your collaborators. In R, three popular style guides are Google’s, Jean Fan’s and the tidyverse’s. The tidyverse’s is very comprehensive and may seem overwhelming at first. You can install the lintr package to automatically check for issues in the styling of your code.
Objects vs. variables
What are known as
objectsinRare known asvariablesin many other programming languages. Depending on the context,objectandvariablecan have drastically different meanings. However, in this lesson, the two words are used synonymously. For more information see: https://cran.r-project.org/doc/manuals/r-release/R-lang.html#Objects
When assigning a value to an object, R does not print anything. You can force R to print the value by using parentheses or by typing the object name:
weight_kg <- 55 # doesn't print anything
(weight_kg <- 55) # but putting parenthesis around the call prints the value of `weight_kg`## [1] 55weight_kg # and so does typing the name of the object## [1] 55Now that R has weight_kg in memory, we can do arithmetic with it. For instance, we may want to convert this weight into pounds (weight in pounds is 2.2 times the weight in kg):
2.2 * weight_kg## [1] 121We can also change an object’s value by assigning it a new one:
weight_kg <- 57.5
2.2 * weight_kg## [1] 126.5This means that assigning a value to one object does not change the values of other objects For example, let’s store the animal’s weight in pounds in a new object, weight_lb:
weight_lb <- 2.2 * weight_kgand then change weight_kg to 100.
weight_kg <- 100What do you think is the current content of the object weight_lb? 126.5 or 220?
Functions and their Arguments
Functions are “canned scripts” that automate more complicated sets of commands including operations assignments, etc. Many functions are predefined, or can be made available by importing R packages (more on that later). A function usually takes one or more inputs called arguments. Functions often (but not always) return a value. A typical example would be the function sqrt(). The input (the argument) must be a number, and the return value (in fact, the output) is the square root of that number. Executing a function (‘running it’) is called calling the function. An example of a function call is:
ten <- sqrt(weight_kg)Here, the value of ten is given to the sqrt() function, the sqrt() function calculates the square root, and returns the value which is then assigned to the object weight_kg. This function is very simple, because it takes just one argument.
The return ‘value’ of a function need not be numerical (like that of sqrt()), and it also does not need to be a single item: it can be a set of things, or even a dataset. We’ll see that when we read data files into R.
Arguments can be anything, not only numbers or filenames, but also other objects. Exactly what each argument means differs per function, and must be looked up in the documentation (see below). Some functions take arguments which may either be specified by the user, or, if left out, take on a default value: these are called options. Options are typically used to alter the way the function operates, such as whether it ignores ‘bad values’, or what symbol to use in a plot. However, if you want something specific, you can specify a value of your choice which will be used instead of the default.
Let’s try a function that can take multiple arguments: round().
round(3.14159)## [1] 3Here, we’ve called round() with just one argument, 3.14159, and it has returned the value 3. That’s because the default is to round to the nearest whole number. If we want more digits we can see how to do that by getting information about the round function. We can use args(round) to find what arguments it takes, or look at the help for this function using ?round.
args(round)## function (x, digits = 0)
## NULL?roundWe see that if we want a different number of digits, we can type digits = 2 or however many we want.
round(3.14159, digits = 2)## [1] 3.14If you provide the arguments in the exact same order as they are defined you don’t have to name them:
round(3.14159, 2)## [1] 3.14And if you do name the arguments, you can switch their order:
round(digits = 2, x = 3.14159)## [1] 3.14It’s good practice to put the non-optional arguments (like the number you’re rounding) first in your function call, and to then specify the names of all optional arguments. If you don’t, someone reading your code might have to look up the definition of a function with unfamiliar arguments to understand what you’re doing.
Vectors and Data Types
A vector is the most common and basic data type in R, and is pretty much the workhorse of R. A vector is composed by a series of values, which can be either numbers or characters. We can assign a series of values to a vector using the c() function. For example we can create a vector of animal weights and assign it to a new object weight_g:
weight_g <- c(50, 60, 65, 82)
weight_g## [1] 50 60 65 82A vector can also contain characters:
animals <- c("mouse", "rat", "dog")
animals## [1] "mouse" "rat" "dog"The quotes around “mouse”, “rat”, etc. are essential here. Without the quotes R will assume objects have been created called mouse, rat and dog. As these objects don’t exist in R’s memory, there will be an error message.
There are many functions that allow you to inspect the content of a vector. length() tells you how many elements are in a particular vector:
length(weight_g)## [1] 4length(animals)## [1] 3An important feature of a vector, is that all of the elements are the same type of data. The function class() indicates the class (the type of element) of an object:
class(weight_g)## [1] "numeric"class(animals)## [1] "character"The function str() provides an overview of the structure of an object and its elements. It is a useful function when working with large and complex objects:
str(weight_g)## num [1:4] 50 60 65 82str(animals)## chr [1:3] "mouse" "rat" "dog"You can use the c() function to add other elements to your vector:
weight_g <- c(weight_g, 90) # add to the end of the vector
weight_g <- c(30, weight_g) # add to the beginning of the vector
weight_g## [1] 30 50 60 65 82 90In the first line, we take the original vector weight_g, add the value 90 to the end of it, and save the result back into weight_g. Then we add the value 30 to the beginning, again saving the result back into weight_g.
We can do this over and over again to grow a vector, or assemble a dataset. As we program, this may be useful to add results that we are collecting or calculating.
An atomic vector is the simplest R data type and is a linear vector of a single type. Above, we saw 2 of the 6 main atomic vector types that R uses: "character" and "numeric" (or "double"). These are the basic building blocks that all R objects are built from. The other 4 atomic vector types are:
"logical"forTRUEandFALSE(the boolean data type)"integer"for integer numbers (e.g.,2L, theLindicates to R that it’s an integer)"complex"to represent complex numbers with real and imaginary parts (e.g.,1 + 4i) and that’s all we’re going to say about them"raw"for bitstreams that we won’t discuss further
You can check the type of your vector using the typeof() function and inputting your vector as the argument.
Vectors are one of the many data structures that R uses. Other important ones are lists (list), matrices (matrix), data frames (data.frame), factors (factor) and arrays (array).
Subsetting vectors
If we want to extract one or several values from a vector, we must provide one or several indices in square brackets. For instance:
animals <- c("mouse", "rat", "dog", "cat")
animals[2]## [1] "rat"animals[c(3, 2)]## [1] "dog" "rat"We can also repeat the indices to create an object with more elements than the original one:
more_animals <- animals[c(1, 2, 3, 2, 1, 4)]
more_animals## [1] "mouse" "rat" "dog" "rat" "mouse" "cat"R indices start at 1. Programming languages like Fortran, MATLAB, Julia, and R start counting at 1, because that’s what human beings typically do. Languages in the C family (including C++, Java, Perl, and Python) count from 0 because that’s simpler for computers to do.
Conditional subsetting
Another common way of subsetting is by using a logical vector. TRUE will select the element with the same index, while FALSE will not:
weight_g <- c(21, 34, 39, 54, 55)
weight_g[c(TRUE, FALSE, FALSE, TRUE, TRUE)]## [1] 21 54 55Typically, these logical vectors are not typed by hand, but are the output of other functions or logical tests. For instance, if you wanted to select only the values above 50:
weight_g > 50 # will return logicals with TRUE for the indices that meet the condition## [1] FALSE FALSE FALSE TRUE TRUE## so we can use this to select only the values above 50
weight_g[weight_g > 50]## [1] 54 55You can combine multiple tests using & (both conditions are true, AND) or | (at least one of the conditions is true, OR):
weight_g[weight_g < 30 | weight_g > 50]## [1] 21 54 55weight_g[weight_g >= 30 & weight_g == 21]## numeric(0)Here, < stands for “less than”, > for “greater than”, >= for “greater than or equal to”, and == for “equal to”. The double equal sign == is a test for numerical equality between the left and right hand sides, and should not be confused with the single = sign, which performs variable assignment (similar to <-).
A common task is to search for certain strings in a vector. One could use the “or” operator | to test for equality to multiple values, but this can quickly become tedious. The function %in% allows you to test if any of the elements of a search vector are found:
animals <- c("mouse", "rat", "dog", "cat")
animals[animals == "cat" | animals == "rat"] # returns both rat and cat## [1] "rat" "cat"animals %in% c("rat", "cat", "dog", "duck", "goat")## [1] FALSE TRUE TRUE TRUEanimals[animals %in% c("rat", "cat", "dog", "duck", "goat")]## [1] "rat" "dog" "cat"Missing data
As R was designed to analyze datasets, it includes the concept of missing data (which is uncommon in other programming languages). Missing data are represented in vectors as NA.
When doing operations on numbers, most functions will return NA if the data you are working with include missing values. This feature makes it harder to overlook the cases where you are dealing with missing data. You can add the argument na.rm = TRUE to calculate the result while ignoring the missing values.
heights <- c(2, 4, 4, NA, 6)
mean(heights)## [1] NAmax(heights)## [1] NAmean(heights, na.rm = TRUE)## [1] 4max(heights, na.rm = TRUE)## [1] 6If your data include missing values, you may want to become familiar with the functions is.na(), na.omit(), and complete.cases(). See below for examples.
## Extract those elements which are not missing values.
heights[!is.na(heights)]## [1] 2 4 4 6## Returns the object with incomplete cases removed. The returned object is an atomic vector of type `"numeric"` (or `"double"`).
na.omit(heights)## [1] 2 4 4 6
## attr(,"na.action")
## [1] 4
## attr(,"class")
## [1] "omit"## Extract those elements which are complete cases. The returned object is an atomic vector of type `"numeric"` (or `"double"`).
heights[complete.cases(heights)]## [1] 2 4 4 6Recall that you can use the typeof() function to find the type of your atomic vector.
Challenge
Using this vector of heights in inches, create a new vector,
heights_no_na, with the NAs removed.heights <- c(63, 69, 60, 65, NA, 68, 61, 70, 61, 59, 64, 69, 63, 63, NA, 72, 65, 64, 70, 63, 65)Use the function
median()to calculate the median of theheightsvector.Use R to figure out how many people in the set are taller than 67 inches.
heights <- c(63, 69, 60, 65, NA, 68, 61, 70, 61, 59, 64, 69, 63, 63, NA, 72, 65, 64, 70, 63, 65) # 1. heights_no_na <- heights[!is.na(heights)] # or heights_no_na <- na.omit(heights) # or heights_no_na <- heights[complete.cases(heights)] # 2. median(heights, na.rm = TRUE)## [1] 64# 3. heights_above_67 <- heights_no_na[heights_no_na > 67] length(heights_above_67)## [1] 6
Now that we have learned how to write scripts, and the basics of R’s data structures, we are ready to start working with the Portal dataset we have been using in the other lessons, and learn about data frames.
Working with Data
Learning Objectives
- Load external data from a .csv file into a data frame.
- Describe what a data frame is.
- Summarize the contents of a data frame.
- Use indexing to subset specific portions of data frames.
- Describe what a factor is.
- Convert between strings and factors.
- Reorder and rename factors.
- Change how character strings are handled in a data frame.
- Format dates.
Presentation of the Survey Data
We are studying the species repartition and weight of animals caught in plots in our study area. The dataset is stored as a comma separated value (CSV) file. Each row holds information for a single animal, and the columns represent:
| Column | Description |
|---|---|
| record_id | Unique id for the observation |
| month | month of observation |
| day | day of observation |
| year | year of observation |
| plot_id | ID of a particular plot |
| species_id | 2-letter code |
| sex | sex of animal (“M”, “F”) |
| hindfoot_length | length of the hindfoot in mm |
| weight | weight of the animal in grams |
| genus | genus of animal |
| species | species of animal |
| taxon | e.g. Rodent, Reptile, Bird, Rabbit |
| plot_type | type of plot |
We are going to use the R function download.file() to download the CSV file that contains the survey data from Figshare, and we will use read.csv() to load into memory the content of the CSV file as an object of class data.frame. Inside the download.file command, the first entry is a character string with the source URL (“https://ndownloader.figshare.com/files/2292169”). This source URL downloads a CSV file from figshare. The text after the comma (“data_raw/portal_data_joined.csv”) is the destination of the file on your local machine. You’ll need to have a folder on your machine called “data_raw” where you’ll download the file. So this command downloads a file from Figshare, names it “portal_data_joined.csv” and adds it to a preexisting folder named “data_raw”.
download.file(url = "https://ndownloader.figshare.com/files/2292169",
destfile = "data_raw/portal_data_joined.csv")You are now ready to load the data:
surveys <- read.csv("data_raw/portal_data_joined.csv")This statement doesn’t produce any output because, as you might recall, assignments don’t display anything. If we want to check that our data has been loaded, we can see the contents of the data frame by typing its name: surveys.
Wow… that was a lot of output. At least it means the data loaded properly. Let’s check the top (the first 6 lines) of this data frame using the function head():
head(surveys)## Try also
View(surveys)Note
read.csvassumes that fields are delineated by commas, however, in several countries, the comma is used as a decimal separator and the semicolon (;) is used as a field delineator. If you want to read in this type of files in R, you can use theread.csv2function. It behaves exactly likeread.csvbut uses different parameters for the decimal and the field separators. If you are working with another format, they can be both specified by the user. Check out the help forread.csv()by typing?read.csvto learn more. There is also theread.delim()for in tab separated data files. It is important to note that all of these functions are actually wrapper functions for the mainread.table()function with different arguments. As such, the surveys data above could have also been loaded by usingread.table()with the separation argument as,. The code is as follows:surveys <- read.table(file="data_raw/portal_data_joined.csv", sep=",", header=TRUE). The header argument has to be set to TRUE to be able to read the headers as by defaultread.table()has the header argument set to FALSE.In addition to the above versions of the csv format, you should develop the habits of looking at and record some parameters of your csv files. For instance, the character encoding, control characters used for line ending, date format (if the date is not splitted into three variables), and the presence of unexpected newlines are important characteristics of your data files. Those parameters will ease up the import step of your data in R.
What are data frames?
Data frames are the de facto data structure for most tabular data, and what we use for statistics and plotting.
A data frame can be created by hand, but most commonly they are generated by the functions read.csv() or read.table(); in other words, when importing spreadsheets from your hard drive (or the web).
A data frame is the representation of data in the format of a table where the columns are vectors that all have the same length. Because columns are vectors, each column must contain a single type of data (e.g., characters, integers, factors). For example, here is a figure depicting a data frame comprising a numeric, a character, and a logical vector.
We can see this when inspecting the structure of a data frame with the function str():
str(surveys)Inspecting data.frame Objects
We already saw how the functions head() and str() can be useful to check the content and the structure of a data frame. Here is a non-exhaustive list of functions to get a sense of the content/structure of the data. Let’s try them out!
- Size:
dim(surveys)- returns a vector with the number of rows in the first element, and the number of columns as the second element (the dimensions of the object)nrow(surveys)- returns the number of rowsncol(surveys)- returns the number of columns
- Content:
head(surveys)- shows the first 6 rowstail(surveys)- shows the last 6 rows
- Names:
names(surveys)- returns the column names (synonym ofcolnames()fordata.frameobjects)rownames(surveys)- returns the row names
- Summary:
str(surveys)- structure of the object and information about the class, length and content of each columnsummary(surveys)- summary statistics for each column Note: most of these functions are “generic”, they can be used on other types of objects besidesdata.frame.
Note: most of these functions are “generic”, they can be used on other types of objects besides data.frame.
## Challenge
## Based on the output of `str(surveys)`, can you answer the following questions?
## * What is the class of the object `surveys`?
## * How many rows and how many columns are in this object?
## * How many species have been recorded during these surveys?Indexing and subsetting data frames
Our survey data frame has rows and columns (it has 2 dimensions), if we want to extract some specific data from it, we need to specify the “coordinates” we want from it. Row numbers come first, followed by column numbers. However, note that different ways of specifying these coordinates lead to results with different classes.
# first element in the first column of the data frame (as a vector)
surveys[1, 1]
# first element in the 6th column (as a vector)
surveys[1, 6]
# first column of the data frame (as a vector)
surveys[, 1]
# first column of the data frame (as a data.frame)
surveys[1]
# first three elements in the 7th column (as a vector)
surveys[1:3, 7]
# the 3rd row of the data frame (as a data.frame)
surveys[3, ]
# equivalent to head_surveys <- head(surveys)
head_surveys <- surveys[1:6, ] : is a special function that creates numeric vectors of integers in increasing or decreasing order, test 1:10 and 10:1 for instance. You can also exclude certain indices of a data frame using the “-” sign:
surveys[, -1] # The whole data frame, except the first column
surveys[-c(7:34786), ] # Equivalent to head(surveys)Data frames can be subset by calling indices (as shown previously), but also by calling their column names directly:
surveys["species_id"] # Result is a data.frame
surveys[, "species_id"] # Result is a vector
surveys[["species_id"]] # Result is a vector
surveys$species_id # Result is a vectorIn RStudio, you can use the autocompletion feature to get the full and correct names of the columns.
Challenge
Create a
data.frame(surveys_200) containing only the data in row 200 of thesurveysdataset.Notice how
nrow()gave you the number of rows in adata.frame?
- Use that number to pull out just that last row in the data frame.
- Compare that with what you see as the last row using
tail()to make sure it’s meeting expectations.- Pull out that last row using
nrow()instead of the row number.- Create a new data frame (
surveys_last) from that last row.Use
nrow()to extract the row that is in the middle of the data frame. Store the content of this row in an object namedsurveys_middle.Combine
nrow()with the-notation above to reproduce the behavior ofhead(surveys), keeping just the first through 6th rows of the surveys dataset.
Factors
When we did str(surveys) we saw that several of the columns consist of integers. The columns genus, species, sex, plot_type, … however, are of a special class called factor. Factors are very useful and actually contribute to making R particularly well suited to working with data. So we are going to spend a little time introducing them. Factors represent categorical data. They are stored as integers associated with labels and they can be ordered or unordered. While factors look (and often behave) like character vectors, they are actually treated as integer vectors by R. So you need to be very careful when treating them as strings. Once created, factors can only contain a pre-defined set of values, known as levels. By default, R always sorts levels in alphabetical order. For instance, if you have a factor with 2 levels:
sex <- factor(c("male", "female", "female", "male"))R will assign 1 to the level "female" and 2 to the level "male" (because f comes before m, even though the first element in this vector is "male"). You can see this by using the function levels() and you can find the number of levels using nlevels():
levels(sex)
nlevels(sex)Sometimes, the order of the factors does not matter, other times you might want to specify the order because it is meaningful (e.g., “low”, “medium”, “high”), it improves your visualization, or it is required by a particular type of analysis. Here, one way to reorder our levels in the sex vector would be:
sex # current order
sex <- factor(sex, levels = c("male", "female"))
sex # after re-orderingIn R’s memory, these factors are represented by integers (1, 2, 3), but are more informative than integers because factors are self describing: "female", "male" is more descriptive than 1, 2. Which one is “male”? You wouldn’t be able to tell just from the integer data. Factors, on the other hand, have this information built in. It is particularly helpful when there are many levels (like the species names in our example dataset).
Using the Tidyverse
Learning Objectives
- Describe the purpose of the
dplyrandtidyrpackages.- Select certain columns in a data frame with the
dplyrfunctionselect.- Select certain rows in a data frame according to filtering conditions with the
dplyrfunctionfilter.- Link the output of one
dplyrfunction to the input of another function with the ‘pipe’ operator%>%.- Add new columns to a data frame that are functions of existing columns with
mutate.- Use the split-apply-combine concept for data analysis.
- Use
summarize,group_by, andcountto split a data frame into groups of observations, apply summary statistics for each group, and then combine the results.- Describe the concept of a wide and a long table format and for which purpose those formats are useful.
- Describe what key-value pairs are.
- Reshape a data frame from long to wide format and back with the
spreadandgathercommands from thetidyrpackage.- Export a data frame to a .csv file.
Data manipulation using dplyr and tidyr
Bracket subsetting is handy, but it can be cumbersome and difficult to read, especially for complicated operations. Enter dplyr. dplyr is a package for making tabular data manipulation easier. It pairs nicely with tidyr which enables you to swiftly convert between different data formats for plotting and analysis.
Packages in R are basically sets of additional functions that let you do more stuff. The functions we’ve been using so far, like str() or data.frame(), come built into R; packages give you access to more of them. Before you use a package for the first time you need to install it on your machine, and then you should import it in every subsequent R session when you need it. You should already have installed the tidyverse package. This is an “umbrella-package” that installs several packages useful for data analysis which work together well such as tidyr, dplyr, ggplot2, tibble, etc.
The tidyverse package tries to address 3 common issues that arise when doing data analysis with some of the functions that come with R:
- The results from a base R function sometimes depend on the type of data.
- Using R expressions in a non standard way, which can be confusing for new learners.
- Hidden arguments, having default operations that new learners are not aware of.
We have seen in our previous lesson that when building or importing a data frame, the columns that contain characters (i.e., text) are coerced (=converted) into the factor data type. We had to set stringsAsFactors to FALSE to avoid this hidden argument to convert our data type.
This time we will use the tidyverse package to read the data and avoid having to set stringsAsFactors to FALSE
If we haven’t already done so, we can type install.packages("tidyverse") straight into the console. In fact, it’s better to write this in the console than in our script for any package, as there’s no need to re-install packages every time we run the script.
Then, to load the package type:
## load the tidyverse packages, incl. dplyr
library(tidyverse)What are dplyr and tidyr?
The package dplyr provides easy tools for the most common data manipulation tasks. It is built to work directly with data frames, with many common tasks optimized by being written in a compiled language (C++). An additional feature is the ability to work directly with data stored in an external database. The benefits of doing this are that the data can be managed natively in a relational database, queries can be conducted on that database, and only the results of the query are returned.
This addresses a common problem with R in that all operations are conducted in-memory and thus the amount of data you can work with is limited by available memory. The database connections essentially remove that limitation in that you can connect to a database of many hundreds of GB, conduct queries on it directly, and pull back into R only what you need for analysis.
The package tidyr addresses the common problem of wanting to reshape your data for plotting and use by different R functions. Sometimes we want data sets where we have one row per measurement. Sometimes we want a data frame where each measurement type has its own column, and rows are instead more aggregated groups - like plots or aquaria. Moving back and forth between these formats is non-trivial, and tidyr gives you tools for this and more sophisticated data manipulation.
To learn more about dplyr and tidyr after the workshop, you may want to check out this handy data transformation with dplyr cheatsheet and this one about tidyr.
We’ll read in our data using the read_csv() function, from the tidyverse package readr, instead of read.csv().
surveys <- read_csv("data_raw/portal_data_joined.csv")You will see the message Parsed with column specification, followed by each column name and its data type. When you execute read_csv on a data file, it looks through the first 1000 rows of each column and guesses the data type for each column as it reads it into R. For example, in this dataset, read_csv reads weight as col_double (a numeric data type), and species as col_character. You have the option to specify the data type for a column manually by using the col_types argument in read_csv.
## inspect the data
str(surveys)## preview the data
View(surveys)Notice that the class of the data is now tbl_df
This is referred to as a “tibble”. Tibbles tweak some of the behaviors of the data frame objects we introduced in the previous episode. The data structure is very similar to a data frame. For our purposes the only differences are that:
- In addition to displaying the data type of each column under its name, it only prints the first few rows of data and only as many columns as fit on one screen.
- Columns of class
characterare never converted into factors.
We’re going to learn some of the most common dplyr functions:
select(): subset columnsfilter(): subset rows on conditionsmutate(): create new columns by using information from other columnsgroup_by()andsummarize(): create summary statistics on grouped dataarrange(): sort resultscount(): count discrete values
Selecting columns and filtering rows
To select columns of a data frame, use select(). The first argument to this function is the data frame (surveys), and the subsequent arguments are the columns to keep.
select(surveys, plot_id, species_id, weight)To select all columns except certain ones, put a “-” in front of the variable to exclude it.
select(surveys, -record_id, -species_id)This will select all the variables in surveys except record_id and species_id.
To choose rows based on a specific criterion, use filter():
filter(surveys, year == 1995)Pipes
What if you want to select and filter at the same time? There are three ways to do this: use intermediate steps, nested functions, or pipes.
With intermediate steps, you create a temporary data frame and use that as input to the next function, like this:
surveys2 <- filter(surveys, weight < 5)
surveys_sml <- select(surveys2, species_id, sex, weight)This is readable, but can clutter up your workspace with lots of objects that you have to name individually. With multiple steps, that can be hard to keep track of.
You can also nest functions (i.e. one function inside of another), like this:
surveys_sml <- select(filter(surveys, weight < 5), species_id, sex, weight)This is handy, but can be difficult to read if too many functions are nested, as R evaluates the expression from the inside out (in this case, filtering, then selecting).
The last option, pipes, are a recent addition to R. Pipes let you take the output of one function and send it directly to the next, which is useful when you need to do many things to the same dataset. Pipes in R look like %>% and are made available via the magrittr package, installed automatically with dplyr. If you use RStudio, you can type the pipe with Ctrl + Shift + M if you have a PC or Cmd + Shift + M if you have a Mac.
surveys %>%
filter(weight < 5) %>%
select(species_id, sex, weight)In the above code, we use the pipe to send the surveys dataset first through filter() to keep rows where weight is less than 5, then through select() to keep only the species_id, sex, and weight columns. Since %>% takes the object on its left and passes it as the first argument to the function on its right, we don’t need to explicitly include the data frame as an argument to the filter() and select() functions any more.
Some may find it helpful to read the pipe like the word “then”. For instance, in the above example, we took the data frame surveys, then we filtered for rows with weight < 5, then we selected columns species_id, sex, and weight. The dplyr functions by themselves are somewhat simple, but by combining them into linear workflows with the pipe, we can accomplish more complex manipulations of data frames.
If we want to create a new object with this smaller version of the data, we can assign it a new name:
surveys_sml <- surveys %>%
filter(weight < 5) %>%
select(species_id, sex, weight)
surveys_smlNote that the final data frame is the leftmost part of this expression.
Mutate
Frequently you’ll want to create new columns based on the values in existing columns, for example to do unit conversions, or to find the ratio of values in two columns. For this we’ll use mutate().
To create a new column of weight in kg:
surveys %>%
mutate(weight_kg = weight / 1000)You can also create a second new column based on the first new column within the same call of mutate():
surveys %>%
mutate(weight_kg = weight / 1000,
weight_lb = weight_kg * 2.2)If this runs off your screen and you just want to see the first few rows, you can use a pipe to view the head() of the data. (Pipes work with non-dplyr functions, too, as long as the dplyr or magrittr package is loaded).
surveys %>%
mutate(weight_kg = weight / 1000) %>%
head()The first few rows of the output are full of NAs, so if we wanted to remove those we could insert a filter() in the chain:
surveys %>%
filter(!is.na(weight)) %>%
mutate(weight_kg = weight / 1000) %>%
head()is.na() is a function that determines whether something is an NA. The ! symbol negates the result, so we’re asking for every row where weight is not an NA.
Split-apply-combine Approach
Many data analysis tasks can be approached using the split-apply-combine paradigm: split the data into groups, apply some analysis to each group, and then combine the results. dplyr makes this very easy through the use of the group_by() function.
The summarize() function
group_by() is often used together with summarize(), which collapses each group into a single-row summary of that group. group_by() takes as arguments the column names that contain the categorical variables for which you want to calculate the summary statistics. So to compute the mean weight by sex:
surveys %>%
group_by(sex) %>%
summarize(mean_weight = mean(weight, na.rm = TRUE))You may also have noticed that the output from these calls doesn’t run off the screen anymore. It’s one of the advantages of tbl_df over data frame.
You can also group by multiple columns:
surveys %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight, na.rm = TRUE)) %>%
tail()Here, we used tail() to look at the last six rows of our summary. Before, we had used head() to look at the first six rows. We can see that the sex column contains NA values because some animals had escaped before their sex and body weights could be determined. The resulting mean_weight column does not contain NA but NaN (which refers to “Not a Number”) because mean() was called on a vector of NA values while at the same time setting na.rm = TRUE. To avoid this, we can remove the missing values for weight before we attempt to calculate the summary statistics on weight. Because the missing values are removed first, we can omit na.rm = TRUE when computing the mean:
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight))Here, again, the output from these calls doesn’t run off the screen anymore. If you want to display more data, you can use the print() function at the end of your chain with the argument n specifying the number of rows to display:
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight)) %>%
print(n = 15)Once the data are grouped, you can also summarize multiple variables at the same time (and not necessarily on the same variable). For instance, we could add a column indicating the minimum weight for each species for each sex:
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight))It is sometimes useful to rearrange the result of a query to inspect the values. For instance, we can sort on min_weight to put the lighter species first:
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight)) %>%
arrange(min_weight)To sort in descending order, we need to add the desc() function. If we want to sort the results by decreasing order of mean weight:
surveys %>%
filter(!is.na(weight)) %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight),
min_weight = min(weight)) %>%
arrange(desc(mean_weight))Counting
When working with data, we often want to know the number of observations found for each factor or combination of factors. For this task, dplyr provides count(). For example, if we wanted to count the number of rows of data for each sex, we would do:
surveys %>%
count(sex) The count() function is shorthand for something we’ve already seen: grouping by a variable, and summarizing it by counting the number of observations in that group. In other words, surveys %>% count() is equivalent to:
surveys %>%
group_by(sex) %>%
summarise(count = n())For convenience, count() provides the sort argument:
surveys %>%
count(sex, sort = TRUE) Previous example shows the use of count() to count the number of rows/observations for one factor (i.e., sex). If we wanted to count combination of factors, such as sex and species, we would specify the first and the second factor as the arguments of count():
surveys %>%
count(sex, species) With the above code, we can proceed with arrange() to sort the table according to a number of criteria so that we have a better comparison. For instance, we might want to arrange the table above in (i) an alphabetical order of the levels of the species and (ii) in descending order of the count:
surveys %>%
count(sex, species) %>%
arrange(species, desc(n))From the table above, we may learn that, for instance, there are 75 observations of the albigula species that are not specified for its sex (i.e. NA).
Exporting data
Now that you have learned how to use dplyr to extract information from or summarize your raw data, you may want to export these new data sets to share them with your collaborators or for archival.
Similar to the read_csv() function used for reading CSV files into R, there is a write_csv() function that generates CSV files from data frames.
Before using write_csv(), we are going to create a new folder, data, in our working directory that will store this generated dataset. We don’t want to write generated datasets in the same directory as our raw data. It’s good practice to keep them separate. The data_raw folder should only contain the raw, unaltered data, and should be left alone to make sure we don’t delete or modify it. In contrast, our script will generate the contents of the data directory, so even if the files it contains are deleted, we can always re-generate them.
In preparation for our next lesson on plotting, we are going to prepare a cleaned up version of the data set that doesn’t include any missing data.
Let’s start by removing observations of animals for which weight and hindfoot_length are missing, or the sex has not been determined:
surveys_complete <- surveys %>%
filter(!is.na(weight), # remove missing weight
!is.na(hindfoot_length), # remove missing hindfoot_length
!is.na(sex)) # remove missing sexBecause we are interested in plotting how species abundances have changed through time, we are also going to remove observations for rare species (i.e., that have been observed less than 50 times). We will do this in two steps: first we are going to create a data set that counts how often each species has been observed, and filter out the rare species; then, we will extract only the observations for these more common species:
## Extract the most common species_id
species_counts <- surveys_complete %>%
count(species_id) %>%
filter(n >= 50)
## Only keep the most common species
surveys_complete <- surveys_complete %>%
filter(species_id %in% species_counts$species_id)### Create the dataset for exporting:
## Start by removing observations for which the `species_id`, `weight`,
## `hindfoot_length`, or `sex` data are missing:
surveys_complete <- surveys %>%
filter(species_id != "", # remove missing species_id
!is.na(weight), # remove missing weight
!is.na(hindfoot_length), # remove missing hindfoot_length
sex != "") # remove missing sex
## Now remove rare species in two steps. First, make a list of species which
## appear at least 50 times in our dataset:
species_counts <- surveys_complete %>%
count(species_id) %>%
filter(n >= 50) %>%
select(species_id)
## Second, keep only those species:
surveys_complete <- surveys_complete %>%
filter(species_id %in% species_counts$species_id)To make sure that everyone has the same data set, check that surveys_complete has nrow(surveys_complete) rows and ncol(surveys_complete) columns by typing dim(surveys_complete).
Now that our data set is ready, we can save it as a CSV file in our data folder.
write_csv(surveys_complete, path = "data/surveys_complete.csv")Elizabeth McDaniel, 2020. License.
Comments
The comment character in R is
#, anything to the right of a#in a script will be ignored by R. It is useful to leave notes and explanations in your scripts. RStudio makes it easy to comment or uncomment a paragraph: after selecting the lines you want to comment, press at the same time on your keyboard Ctrl + Shift + C. If you only want to comment out one line, you can put the cursor at any location of that line (i.e. no need to select the whole line), then press Ctrl + Shift + C.